To work with a database in dplyr, you must first connect to it, using DBI::dbConnect. We’re not going to go into the details of the DBI package here, but it’s the foundation upon which dbplyr is built. You’ll need to learn more about if you need to do things to the database that are beyond the scope of dplyr. Overview dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges: mutate adds new variables that are functions of existing variables select picks variables based on their names. Practice Problems using dplyr 5 ) Onto a new example: Calculate the total births in each maternal race/ethnicity group (WnH, AA, WH, AI/AN, Other). For each group, also calculate the prevalence of early care and prevalence of preterm birth. When calculating nested counts use dplyr::vars to specify 2 variables for targetvar. Setdistinctby(e, distinctby) - Specifies variable(s) to use to calculate distinct occurrences. DISTINCT VS EVENT COUNTS FORMATTING setformatstrings and fstr are used to specify the occurrence and proportion variables and how they will be presented.
Complete List of Cheat Sheets and Infographics for Artificial intelligence (AI), Neural Networks, Machine Learning, Deep Learning and Big Data.
Content Summary
Neural Networks
Neural Networks Graphs
Machine Learning Overview
Machine Learning: Scikit-learn algorithm
Scikit-Learn
Machine Learning: Algorithm Cheat Sheet
Python for Data Science
TensorFlow
Keras
Numpy
Pandas
Data Wrangling
Data Wrangling with dplyr and tidyr
Scipy
Matplotlib
Data Visualization
PySpark
Big-O
Resources
Neural Networks
Artificial neural networks (ANN) or connectionist systems are computing systems vaguely inspired by the biological neural networks that constitute animal brains. The neural network itself is not an algorithm, but rather a framework for many different machine learning algorithms to work together and process complex data inputs. Such systems “learn” to perform tasks by considering examples, generally without being programmed with any task-specific rules.
Neural Networks Graphs
Graph Neural Networks (GNNs) for representation learning of graphs broadly follow a neighborhood aggregation framework, where the representation vector of a node is computed by recursively aggregating and transforming feature vectors of its neighboring nodes. Many GNN variants have been proposed and have achieved state-of-the-art results on both node and graph classification tasks.
Machine Learning Overview
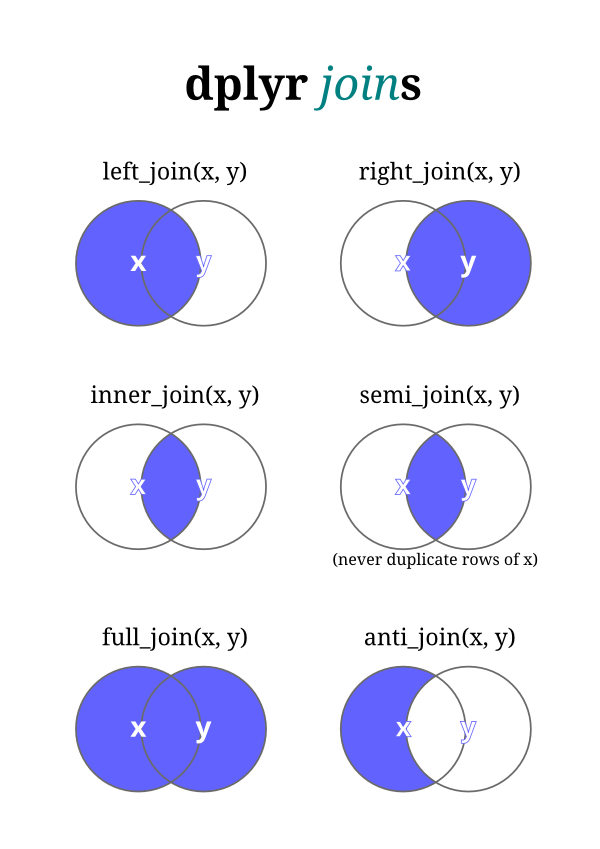
Rstudio Dplyr Cheat Sheet
Machine learning (ML) is the scientific study of algorithms and statistical models that computer systems use to progressively improve their performance on a specific task. Machine learning algorithms build a mathematical model of sample data, known as “training data”, in order to make predictions or decisions without being explicitly programmed to perform the task. Machine learning algorithms are used in the applications of email filtering, detection of network intruders, and computer vision, where it is infeasible to develop an algorithm of specific instructions for performing the task.
Machine Learning: Scikit-learn algorithm
This machine learning cheat sheet will help you find the right estimator for the job which is the most difficult part. The flowchart will help you check the documentation and rough guide of each estimator that will help you to know more about the problems and how to solve it.
Scikit-Learn
Scikit-learn (formerly scikits.learn) is a free software machine learning library for the Python programming language. It features various classification, regression and clustering algorithms including support vector machines, random forests, gradient boosting, k-means and DBSCAN, and is designed to interoperate with the Python numerical and scientific libraries NumPy and SciPy.
Machine Learning: Algorithm Cheat Sheet
This machine learning cheat sheet from Microsoft Azure will help you choose the appropriate machine learning algorithms for your predictive analytics solution. First, the cheat sheet will asks you about the data nature and then suggests the best algorithm for the job.
Python for Data Science
TensorFlow
In May 2017 Google announced the second-generation of the TPU, as well as the availability of the TPUs in Google Compute Engine. The second-generation TPUs deliver up to 180 teraflops of performance, and when organized into clusters of 64 TPUs provide up to 11.5 petaflops.
Keras
In 2017, Google’s TensorFlow team decided to support Keras in TensorFlow’s core library. Chollet explained that Keras was conceived to be an interface rather than an end-to-end machine-learning framework. It presents a higher-level, more intuitive set of abstractions that make it easy to configure neural networks regardless of the backend scientific computing library.
Numpy
NumPy targets the CPython reference implementation of Python, which is a non-optimizing bytecode interpreter. Mathematical algorithms written for this version of Python often run much slower than compiled equivalents. NumPy address the slowness problem partly by providing multidimensional arrays and functions and operators that operate efficiently on arrays, requiring rewriting some code, mostly inner loops using NumPy.
Pandas
The name ‘Pandas’ is derived from the term “panel data”, an econometrics term for multidimensional structured data sets.
Data Wrangling
The term “data wrangler” is starting to infiltrate pop culture. In the 2017 movie Kong: Skull Island, one of the characters, played by actor Marc Evan Jackson is introduced as “Steve Woodward, our data wrangler”.
Data Wrangling with dplyr and tidyr
Scipy
SciPy builds on the NumPy array object and is part of the NumPy stack which includes tools like Matplotlib, pandas and SymPy, and an expanding set of scientific computing libraries. This NumPy stack has similar users to other applications such as MATLAB, GNU Octave, and Scilab. The NumPy stack is also sometimes referred to as the SciPy stack.
Matplotlib
matplotlib is a plotting library for the Python programming language and its numerical mathematics extension NumPy. It provides an object-oriented API for embedding plots into applications using general-purpose GUI toolkits like Tkinter, wxPython, Qt, or GTK+. There is also a procedural “pylab” interface based on a state machine (like OpenGL), designed to closely resemble that of MATLAB, though its use is discouraged. SciPy makes use of matplotlib. pyplot is a matplotlib module which provides a MATLAB-like interface. matplotlib is designed to be as usable as MATLAB, with the ability to use Python, with the advantage that it is free.
Data Visualization
PySpark
Big-O
Big O notation is a mathematical notation that describes the limiting behavior of a function when the argument tends towards a particular value or infinity. It is a member of a family of notations invented by Paul Bachmann, Edmund Landau and others, collectively called Bachmann–Landau notation or asymptotic notation.
Resources
Big-O Algorithm Cheat Sheet
Bokeh Cheat Sheet
Data Science Cheat Sheet
Data Wrangling Cheat Sheet
Data Wrangling
Ggplot Cheat Sheet
Keras Cheat Sheet
Keras
Machine Learning Cheat Sheet
Machine Learning Cheat Sheet
ML Cheat Sheet
Matplotlib Cheat Sheet
Matpotlib
Neural Networks Cheat Sheet
Neural Networks Graph Cheat Sheet
Neural Networks
Numpy Cheat Sheet
NumPy
Pandas Cheat Sheet
Pandas
Pandas Cheat Sheet
Pyspark Cheat Sheet
Scikit Cheat Sheet
Scikit-learn
Scikit-learn Cheat Sheet
Scipy Cheat Sheet
SciPy
TesorFlow Cheat Sheet
Tensor Flow
Course Duck > The World’s Best Machine Learning Courses & Tutorials in 2020
Tag: Machine Learning, Deep Learning, Artificial Intelligence, Neural Networks, Big Data
Related posts:
As well as working with local in-memory data stored in data frames, dplyr also works with remote on-disk data stored in databases. This is particularly useful in two scenarios:
Your data is already in a database.
You have so much data that it does not all fit into memory simultaneouslyand you need to use some external storage engine.
(If your data fits in memory, there is no advantage to putting it in a database; it will only be slower and more frustrating.)
This vignette focuses on the first scenario because it is the most common. If you are using R to do data analysis inside a company, most of the data you need probably already lives in a database (it’s just a matter of figuring out which one!). However, you will learn how to load data in to a local database in order to demonstrate dplyr’s database tools. At the end, I’ll also give you a few pointers if you do need to set up your own database.
Getting started
To use databases with dplyr, you need to first install dbplyr:
You’ll also need to install a DBI backend package. The DBI package provides a common interface that allows dplyr to work with many different databases using the same code. DBI is automatically installed with dbplyr, but you need to install a specific backend for the database that you want to connect to.
Five commonly used backends are:
RMySQLconnects to MySQL and MariaDB
RPostgreSQLconnects to Postgres and Redshift.
RSQLite embeds a SQLite database.
odbc connects to many commercialdatabases via the open database connectivity protocol.
bigrquery connects to Google’sBigQuery.
If the database you need to connect to is not listed here, you’ll need to do some investigation yourself.
In this vignette, we’re going to use the RSQLite backend, which is automatically installed when you install dbplyr. SQLite is a great way to get started with databases because it’s completely embedded inside an R package. Unlike most other systems, you don’t need to set up a separate database server. SQLite is great for demos, but is surprisingly powerful, and with a little practice you can use it to easily work with many gigabytes of data.
Connecting to the database
To work with a database in dplyr, you must first connect to it, using DBI::dbConnect(). We’re not going to go into the details of the DBI package here, but it’s the foundation upon which dbplyr is built. You’ll need to learn more about if you need to do things to the database that are beyond the scope of dplyr.
The arguments to DBI::dbConnect() vary from database to database, but the first argument is always the database backend. It’s RSQLite::SQLite() for RSQLite, RMySQL::MySQL() for RMySQL, RPostgreSQL::PostgreSQL() for RPostgreSQL, odbc::odbc() for odbc, and bigrquery::bigquery() for BigQuery. SQLite only needs one other argument: the path to the database. Here we use the special string, ':memory:', which causes SQLite to make a temporary in-memory database.
Most existing databases don’t live in a file, but instead live on another server. In real life that your code will look more like this:
(If you’re not using RStudio, you’ll need some other way to securely retrieve your password. You should never record it in your analysis scripts or type it into the console.)
Our temporary database has no data in it, so we’ll start by copying over nycflights13::flights using the convenient copy_to() function. This is a quick and dirty way of getting data into a database and is useful primarily for demos and other small jobs.
As you can see, the copy_to() operation has an additional argument that allows you to supply indexes for the table. Here we set up indexes that will allow us to quickly process the data by day, carrier, plane, and destination. Creating the write indices is key to good database performance, but is unfortunately beyond the scope of this article.

Now that we’ve copied the data, we can use tbl() to take a reference to it:
When you print it out, you’ll notice that it mostly looks like a regular tibble:
The main difference is that you can see that it’s a remote source in a SQLite database.
Generating queries
To interact with a database you usually use SQL, the Structured Query Language. SQL is over 40 years old, and is used by pretty much every database in existence. The goal of dbplyr is to automatically generate SQL for you so that you’re not forced to use it. However, SQL is a very large language, and dbplyr doesn’t do everything. It focuses on SELECT statements, the SQL you write most often as an analyst.
Most of the time you don’t need to know anything about SQL, and you can continue to use the dplyr verbs that you’re already familiar with:

However, in the long run, I highly recommend you at least learn the basics of SQL. It’s a valuable skill for any data scientist, and it will help you debug problems if you run into problems with dplyr’s automatic translation. If you’re completely new to SQL, you might start with this codeacademy tutorial. If you have some familiarity with SQL and you’d like to learn more, I found how indexes work in SQLite and 10 easy steps to a complete understanding of SQL to be particularly helpful.
The most important difference between ordinary data frames and remote database queries is that your R code is translated into SQL and executed in the database, not in R. When working with databases, dplyr tries to be as lazy as possible:
It never pulls data into R unless you explicitly ask for it.
It delays doing any work until the last possible moment: it collects togethereverything you want to do and then sends it to the database in one step.
For example, take the following code:
Surprisingly, this sequence of operations never touches the database. It’s not until you ask for the data (e.g., by printing tailnum_delay) that dplyr generates the SQL and requests the results from the database. Even then it tries to do as little work as possible and only pulls down a few rows.
Behind the scenes, dplyr is translating your R code into SQL. You can see the SQL it’s generating with show_query():
If you’re familiar with SQL, this probably isn’t exactly what you’d write by hand, but it does the job. You can learn more about the SQL translation in vignette('sql-translation').
Typically, you’ll iterate a few times before you figure out what data you need from the database. Once you’ve figured it out, use collect() to pull all the data down into a local tibble:
collect() requires that database does some work, so it may take a long time to complete. Otherwise, dplyr tries to prevent you from accidentally performing expensive query operations:
Because there’s generally no way to determine how many rows a query willreturn unless you actually run it,
nrow()is alwaysNA.Because you can’t find the last few rows without executing the wholequery, you can’t use
tail().
You can also ask the database how it plans to execute the query with explain(). The output is database-dependent and can be esoteric, but learning a bit about it can be very useful because it helps you understand if the database can execute the query efficiently, or if you need to create new indices.

Creating your own database
If you don’t already have a database, here’s some advice from my experiences setting up and running all of them. SQLite is by far the easiest to get started with, but the lack of window functions makes it limited for data analysis. PostgreSQL is not too much harder to use and has a wide range of built-in functions. In my opinion, you shouldn’t bother with MySQL/MariaDB; it’s a pain to set up, the documentation is sub par, and it’s less feature-rich than Postgres. Google BigQuery might be a good fit if you have very large data, or if you’re willing to pay (a small amount of) money to someone who’ll look after your database.
All of these databases follow a client-server model - a computer that connects to the database and the computer that is running the database (the two may be one and the same, but usually aren’t). Getting one of these databases up and running is beyond the scope of this article, but there are plenty of tutorials available on the web.
MySQL/MariaDB
In terms of functionality, MySQL lies somewhere between SQLite and PostgreSQL. It provides a wider range of built-in functions, but it does not support window functions (so you can’t do grouped mutates and filters).
PostgreSQL
PostgreSQL is a considerably more powerful database than SQLite. It has:
Dplyr Cheat Sheet 2020
a much wider range of built-in functions, and
support for window functions, which allow grouped subset and mutates to work.
BigQuery
BigQuery is a hosted database server provided by Google. To connect, you need to provide your project, dataset and optionally a project for billing (if billing for project isn’t enabled).
It provides a similar set of functions to Postgres and is designed specifically for analytic workflows. Because it’s a hosted solution, there’s no setup involved, but if you have a lot of data, getting it to Google can be an ordeal (especially because upload support from R is not great currently). (If you have lots of data, you can ship hard drives!)
